The CARRE conceptual model of the risk factor led to the development of a novel ontology for risk factors in medicine. The ontology was developed using natural language tools for knowledge representation. In particular, we used the OWL Simplified Englishontology editor developed during the SWAT project, and presented an automatically generated textual glossary version of the ontology from the SWAT Tools verbaliser. This is exported in OWL/XML as a final stage. External vocabularies are not directly referenced within the ontology; rather, the appropriate mappings between CARRE terms and external terms are represented in an external file in the Turtle format. This approach allows flexibility and maintainability without sacrificing interoperability or expressiveness.
The “core” of the CARRE ontology is the semantic representation of the CARRE data model for risk (D.2.2), intended to represent current medical knowledge regarding cardiorenal risk factors. This core relates to public data; that is to say, data from the medical literature regarding conditions, genetics, demographics and the environment, how these factors interact and the studies and evidence quality relating to their interactions.
CARRE ontology links to a number of relevant external standardized vocabularies, namely: ICD9/10 and SNOMED-CT (for diseases), ICD10PCS (for interventions), FOAF and HL7 (for demographics), UMLS (for genetic, environmental and behavioural concepts), W3C ontologies for time, time zone and geography, CMO and LOINC (for measurements), and QUDT and UO (for units of measurement).
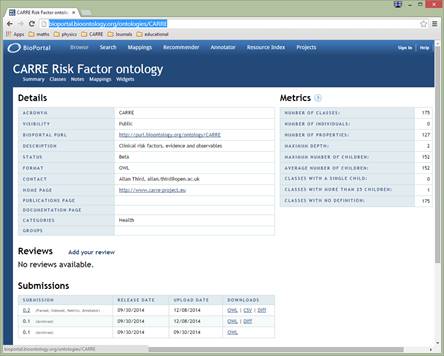
CARRE ontology is published in the NCBO Bioportal, (at http://bioportal.bioontology.org/ontologies/CARRE)
Author: Allan Third, George Gkotsis (OU)
Date: 25 September 2014